28Next generation sequencing data assembly
Next generation sequencing data assembly
BioNumerics’ Power Assembler is designed for pre-processing and assembly of Next Generation Sequencing (NGS) data. The Power Assembler contains powerful algorithms for both mapped assembly using one or multiple reference sequences (re-sequencing) and de novo assembly into contigs of up to full genome size from millions of reads. This versatile sequence assembly tool accepts data from Roche 454, Illumina Solexa and FASTA or FASTQ files (Applied Biosystems SOLiD, Ion Torrent). With the Power Assembler, the user can build pipelines that contain a specific series of steps (“actions”) in the pre-processing and template-assisted assembly of high throughput sequencing projects.
Typical actions include:
· Loading reads (+ quality info).
· Loading reference sequence(s).
· Demultiplexing.
· Paired end splitting.
· Trimming and filtering of reads according to various quality criteria.
· Calculating global statistics on the project.
· Aligning the reads against the reference sequence(s).
· Exporting the sequence(s) to the BioNumerics database.
Through the availability of a large number of predefined actions, the user can easily set up a basic assembly project pipeline. However, more experienced users can break down each action into its atomic operators, and edit the pipeline at the deepest level or construct custom actions. The degree of flexibility with which workflows can thus be generated is practically unlimited. Action pipelines can be saved as templates and shared among users.
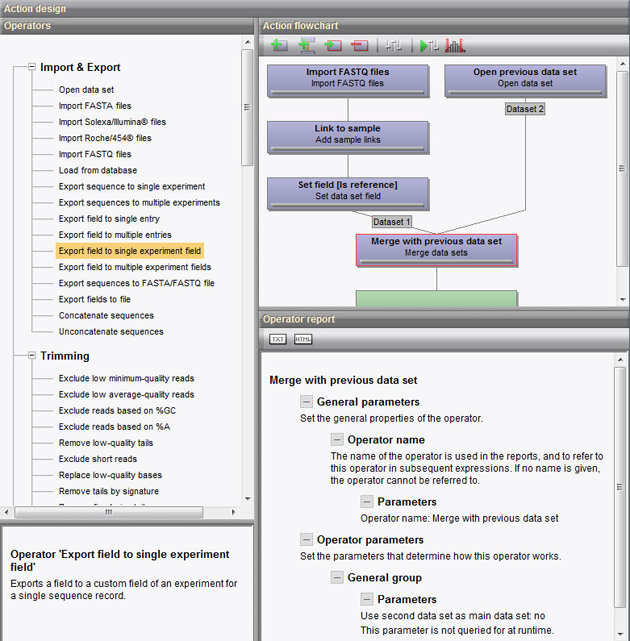
The interface is fully graphical with surveyable flowcharts for pipelines and actions and allows detailed reports, graphs and statistics to be calculated for each component. The Summary graphs resulting from specific actions (e.g. quality trimming, coverage histogram) allow parameters such as minimum read quality and read length to be set directly on the histogram.
Action pipelines can be saved as templates which can be used for automated high throughput pre-processing and analysis. Depending on the type of project, pipelines can be tuned to run fully automatically, with all parameters predefined, or can request user input at specific steps. Pipeline templates can be modified and saved into new templates if requested.
Power Assembler projects can be opened from any other sequence analysis window in the BioNumerics software, including the sequence editor, the comparison window, the multiple alignment window, the chromosome comparison window, etc. The viewport in the Power Assembler is automatically adjusted to the selected base in the analysis window so that doubtful bases or important SNPs can easily be verified from the source.
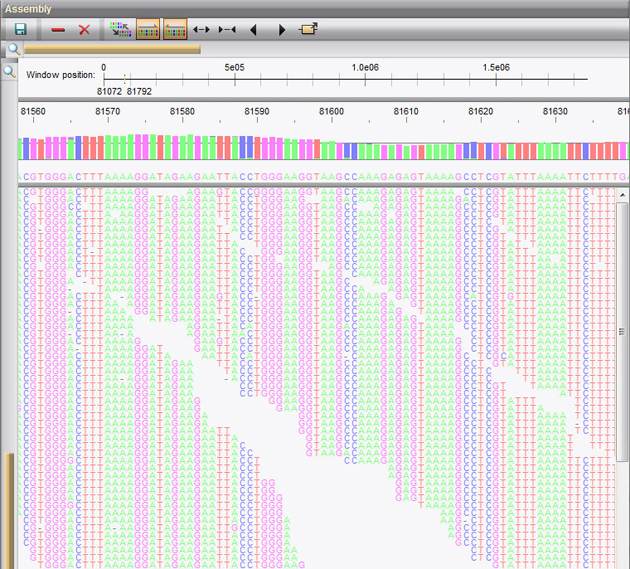
In combination with the Chromosome comparison tools and the Annotation tools, the Power Assembler renders BioNumerics into a powerful and integrated chromosome analysis platform.
Required modules:
