33Primer design
Primer design
The PCR primer design tool in the BioNumerics software searches for optimal primers or primer combinations for the most diverse experiment setups by taking into account a large number of experimental parameters. The user can specify various primer properties such as preferential length and melting temperature, %GC boundaries, maximal degeneracy, etc. Forward and reverse primers and PCR combinations can be sorted and selected according to different parameters (position, melting temperature, length, degeneracy, %GC...). Selected primer pairs can be stored in the oligonucleotide database.
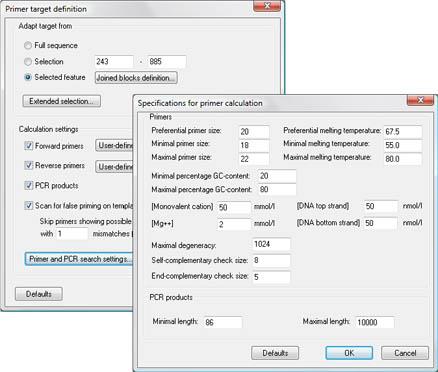
In combination with the Tree and Network Inference module, primer designing becomes possible in the Multiple alignment window. This allows you to identify primer pairs that discriminate between defined groups of sequences, e.g. for application in highly specific PCR detection assays.
Required modules:
See tutorial:
